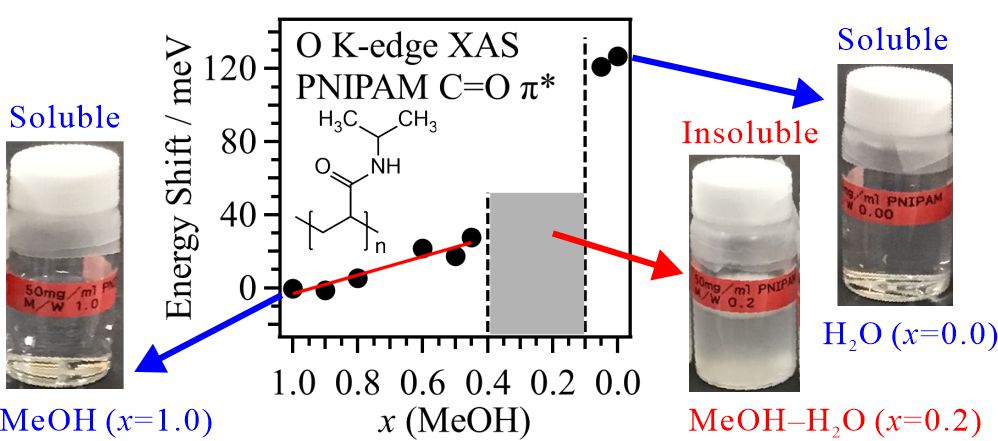
The XAS measurement was applied to polymers in solutions. The cononsolvency mechanism of poly(N-isopropylacrylamide) (PNIPAM), dissolving in pure methanol (MeOH) and water but being insoluble in aqueous MeOH solutions, were investigated by O K-edge XAS [1]. Figure 1 shows the energy shift of the C=O π* peaks in PNIPAM as a function of MeOH molar fraction at 25 °C. In the MeOH-rich region, the energy shifts of the C=O π* peaks are higher in the mixed solvent than in pure MeOH. In contrast, the energy shift of the C=O π* peak of PNIPAM is much higher in pure H2O than in pure MeOH. Although the dissolution behaviors of PNIPAM in H2O and MeOH are identical on the macroscopic scale, the molecular interactions of PNIPAM with H2O and MeOH are very different on the microscopic scale.
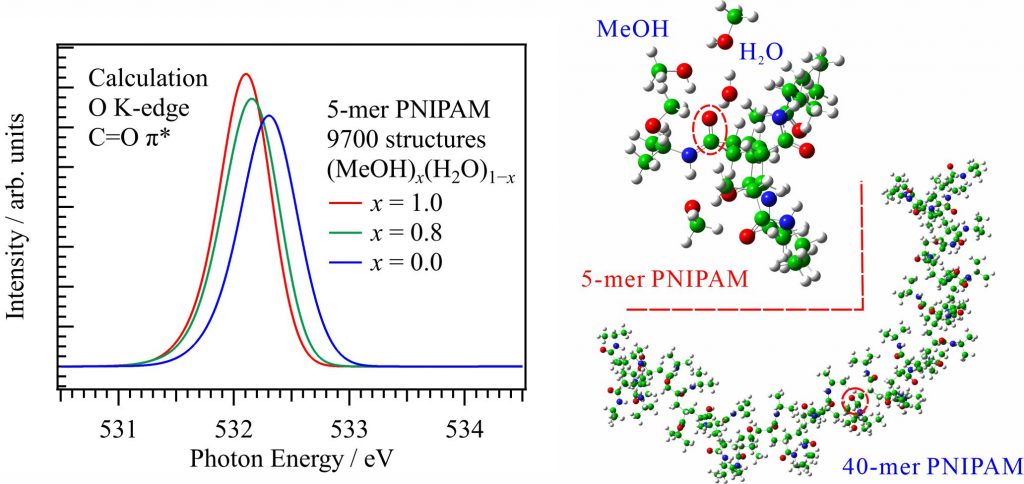
For obtaining the O K-edge inner-shell spectra of PNIPAM in solutions, the 5-mer PNIPAM chains with terminated H atoms, including the second coordination shells of the solvent methanol and water molecules, were extracted from the 40-mer PNIPAM chains in solutions, whose snapshots were obtained by the molecular dynamics simulations [2]. The inner-shell spectra of PNIPAM in aqueous methanol solutions shown in Fig. 2 were obtained by averaging those of 9700 extracted polymer structures. This calculation method can be precisely evaluated the energy shifts of the C=O π* peaks of PNIPAM caused by the structural changes of the polymer chains, the substitutions of the hydrogen bonds of the C=O groups in PNIPAM from methanol to water molecules, and the increases of the coordination numbers of solvent molecules with the C=O groups, which were observed in the O K-edge XAS experiments.